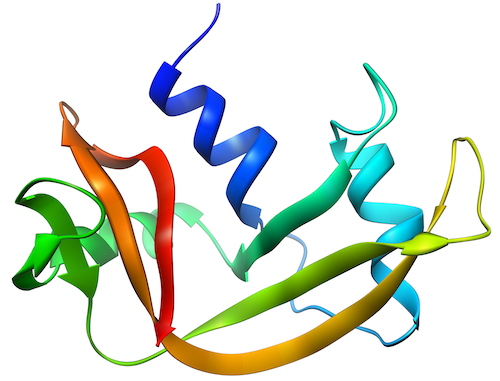
Right as we were in the middle of moving last year, I helped out with the data processing of an interesting experiment (see here and here). The goal of the lead researcher, Tim Ryan, was to get proteins to change morphology upon application of pressure and elevated temperature. The publication on that came out a few months ago, and is available here.
It was found that the application of temperature alone could denature the protein, but applying only elevated pressure (up to about 4.5 kBar) was insufficient to effect unfolding (literature indicates that this would occur beyond about 6 kBar for this protein). However, the combination of pressure and temperature brings about a time-dependent unfolding of the protein, culminating in a structure which differs from that obtained in the temperature-only denaturation.
As for the remote simultaneous data corrections, they were fairly streamlined this time around. The processed data was largely available during the beamtime already, which helped the team make decisions on what to focus on in the final measurements. Therefore, I’m quite happy with the result, and my personal goal was achieved.

Leave a Reply