Since the last publication on McSAS, we’ve been working hard on improving the structure and features (i.e. invisible and visible improvements). This beta version is already available from the Git repository as a Python program. The biggest changes are as follows:
A whole new tab
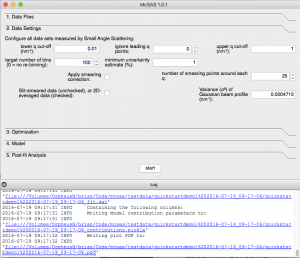
A “Data settings”-tab is available in the GUI, allowing the modification of the data properties (Figure 1). In here, you can clip the lower- and upper limit of the data. For the lower limit, you can clip either by q-value, or by the amount of datapoints to ignore.
Rebinning
Rebinning has some advantages, not in the least the improvement of uncertainty estimates and computation time. Therefore, rebinning has been set to (by default) rebin your data to 100 datapoints on a logarithmic Q-scale. This is only a target value, so depending on your data you might end up with slightly fewer datapoints than the set-point (for example, when bins with no datapoints are removed).
Smearing options
We have also included smearing in the data settings, to account for pinhole smearing effects and slit smearing effects. This is implemented and tested, but the user interface for this will change somewhat. User-supplied smearing profiles may be possible in a future upgrade, but only when using the new NXCanSAS data format.
Cumulative distribution function
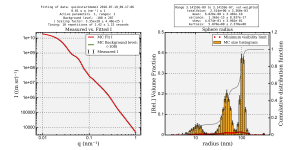
We now show a cumulative distribution function as a grey line in the size distribution plot (Figure 2). This way, you can easily determine parameters such as the D10, D50 and D90 values.
Advanced settings
Advanced settings are available in the Optimization tab, allowing the advanced user to change, amongst others, the number of contributions, the weighting compensation factor (works best between 0.5 and 0.7), and whether or not to fit a flat background factor.
Further work
Some of the things we are working on at the moment, include the implementation of an API for easier integration of McSAS in existing data reduction schemes. Besides that, it should become possible for users to write and include their own models in the program. Finally, there is a good chance we will get multicore processing working cross-platform for a decent speed-boost!
None of this would have been possible without the herculean effort by Ingo Bressler. Many thanks to him for his work!

Leave a Reply