
[update: linux versions (32-bit and 64-bit) are now also available. The article has been changed to reflect that.]
We are very happy to be able to introduce McSAS v1.3.0 as compiled binaries for Windows, os X, and Linux! Thanks to the continuous efforts of Ingo Bressler, the new version introduces a slew of updates and improvements. I’ll highlight the most interesting ones:
First and foremost, there have been many, many updates in the underlying code. This makes it more stable for many applications, and more flexible for us. For example, the way data is handled underneath is now a lot more flexible, travelling in a nice class of its own. These updates are invisible, yet bring with it structural improvements, and should not be underestimated. As an added bonus, the underlying program code is now also compatible with Python3.
Externally visible improvements include:
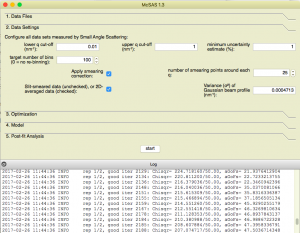
- The ability to limit the data fitting range (a minimum- and maximum Q can be set)
- Automatic rebinning of the data to log-spaced datapoints, with error propagation. The number of target datapoints can be set, the actual number of binned datapoints is this number or slightly less: empty binned datapoints are skipped.
- Basic (forward) smearing compensation for pinhole-collimated systems is implemented, for increased precision. For this, the width
of a gaussian approximation to the beam profile must be provided. Slit-smearing compensation will follow in v1.3.1, allowing you to fit slit-smeared data instead of the more noisy de-smeared data.
- You can use self-written model functions to fit, these will be automatically found if they are placed together with the other fitting models in the program directory. This is “[program directory]/models” under windows, or “[mcsas package name]/Contents/MacOS/models” under os X (right-click the McSAS-1.3 app, and select “show package contents” to get there).
- The entire program set-up is stored in a hierarchical structure inside an HDF5 container. It is NeXus compatible in the sense that it sits in a separate hierarchy, which can live alongside the NeXus hierarchy. In the future, it is intended that this forms the API for external set-up of McSAS optimisations.
- The series development of integral parameters can be shown, giving you a good insight into the systematic changes of an in-situ-monitored experiment.
- An “alternative to the Goodness-of-Fit (aGoF)” is also shown. As highlighted in this paper, it is a separate measure that can indicate poor uncertainty estimates on the data (this would be the case if the aGoF value differs significantly from the GoF measure).
So grab yourself a copy from the downloads section of mcsas.net, and let us know what you think!

Leave a Reply