One persistent problem in the X-ray scattering community is the lack of data interoperability. This is exactly what kicked off the CanSAS collaboration 40 years ago, with the initial intention to make a translator between different formats.
With the push to HDF5-based archival storage formats (ideally using the NeXus/NXsas recommended structure for raw data and NXcanSAS for processed data), a lot of the problems of storing and reading information were solved. Unfortunately, the world isn’t as consistently using NXsas and NXcanSAS as we might hope, and what our various instruments produce can be far off from what we need. So, together with Glen Smales, Abdul Moeez, Ingo Breßler, and Anja Hörmann, we have started working on a universal solution…
Why would you need this?
Imagine you are searching for a way to, for example, use our universal data correction pipeline on your X-ray scattering data to automatically and reliably get trustworthy data. Or you have data from an electron microscope or tomography instrument and would like to turn this into information into something better structured, ready for automated processing and cataloguing in databases. You need to transform the files from the machine into something sensible.
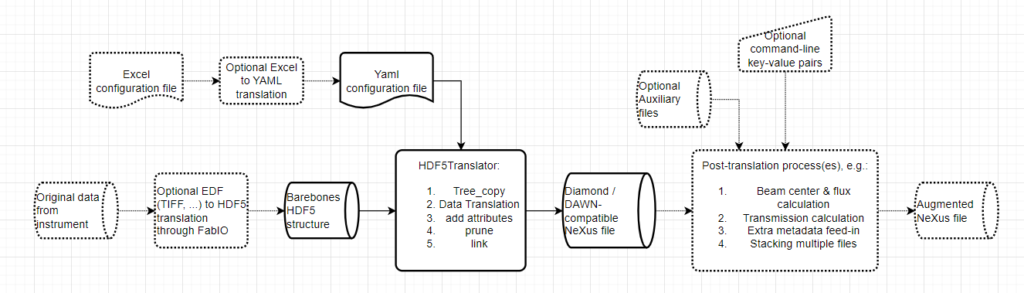
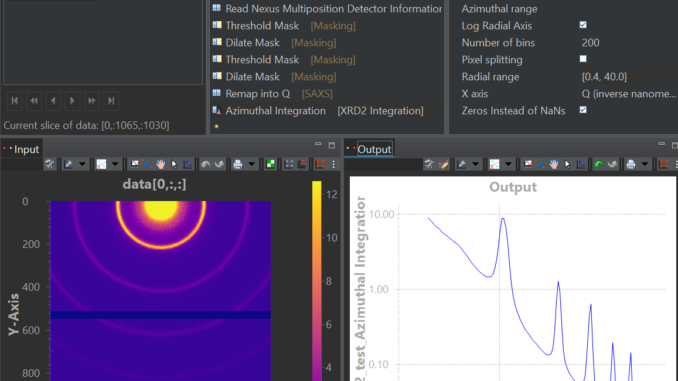
This is where HDF5Translator comes into play (Figure 1). It is a core component of any data preprocessing strategy: it can read information from one file, apply validation, translation and/or conversion rules, and put it in another location in the output file. You can use it once or mutliple times to mix in more files, and tack on additional, more complex post-translation processes such as beam analyses or stacking operations (a framework for which is also supplied). The output can be easily made compatible with Diamond’s open-source DAWN’s processing framework, so you can immediately start building your correction pipeline or adapt one of ours (Figure 2, 3). Three extensive translation examples are available already in our associated library available on Zenodo.
What it does in detail
(Skip this section if you’re not interested in the nitty gritty…)
HDF5Translator does a couple of things sequentially, in an attempt to make a “smart” conversion. The following steps are done in sequence:
- Copy the template into the output file location
- Copy entire (sub-)trees from the input to the output file unchanged. This behaves like rsync when it comes to trailing slashes, so you have to be precise in your definition.
- Translate particular datasets from a location in the source to a new location in the destination. More specifically, this means in sequence:
- adjust the data for the destination: UTF-8-decode strings, find out if strings represent an array, cast data into the target datatype
- Apply the optional user-supplied (lambda) transformation to the data. These can be very powerful, the BAM example shows how to use these to add timestamps and average over data. Note that the inclusion of lambda functions offers extreme translation flexibility, but also allows for arbitrary command execution. Make sure to containerize the translator with correct permissions, if trust of its users is not a given.
- Perform the unit conversion to the target units using the Pint library (if any units are specified in the source or configuration, they are added to the attributes). Additional units of “eigerpixel” and “pilatuspixel” have also been defined (in units of length) to convert from pixel to mm or vice versa
- Add 1-sized dimensions if needed to meet the minimum required dimensionality setting
- Turn arrays of strings into plain strings (Anton Paar stores any string as an array of strings, which can be a problem upon reading)
- Add any additional specified attributes, sanitizing the attributes before storage to string, float or array
- Update attributes of groups and datasets according to the attributes sheet in the excel file
- Remove (prune) any unwanted tree entries
- Create internal or external links between datasets and groups. An alternate source file (with optional wildcards!) can be provided if you want to link to data in an external file.
Additional tools
If your initial files are not in HDF5 but in, say, legacy formats like EDF, you can use the supplied “edf_to_hdf5.py” tool (internally using fabIO) to translate it into a flat HDF5 file ready for translation. This tool can be extended or modified to work with other storage formats easily. Translation configurations are supplied as yaml files, but you can translate an easier-to-use excel sheet version into yaml using the supplied “excel_translator.py” tool. We will talk about post-translation operations below.
A library of usage examples
To show that HDF5Translator can be useful for you, we are also supplying a separate library of usage examples. At the moment, this just has three examples, which also might need a little more fine-tuning, but we hope this will grow as more people start participating. The examples naturally include the MOUSE, but also the University of Washington’s Xenocs Xeuss 3.0 instrument, and the TU Graz’ Anton Paar SAXSPoint2 instrument. The README.md files in each example will tell you what to do to get this conversion going. Note that these translations may be updated or extended in the future to improve the structure or include more auxiliary data.
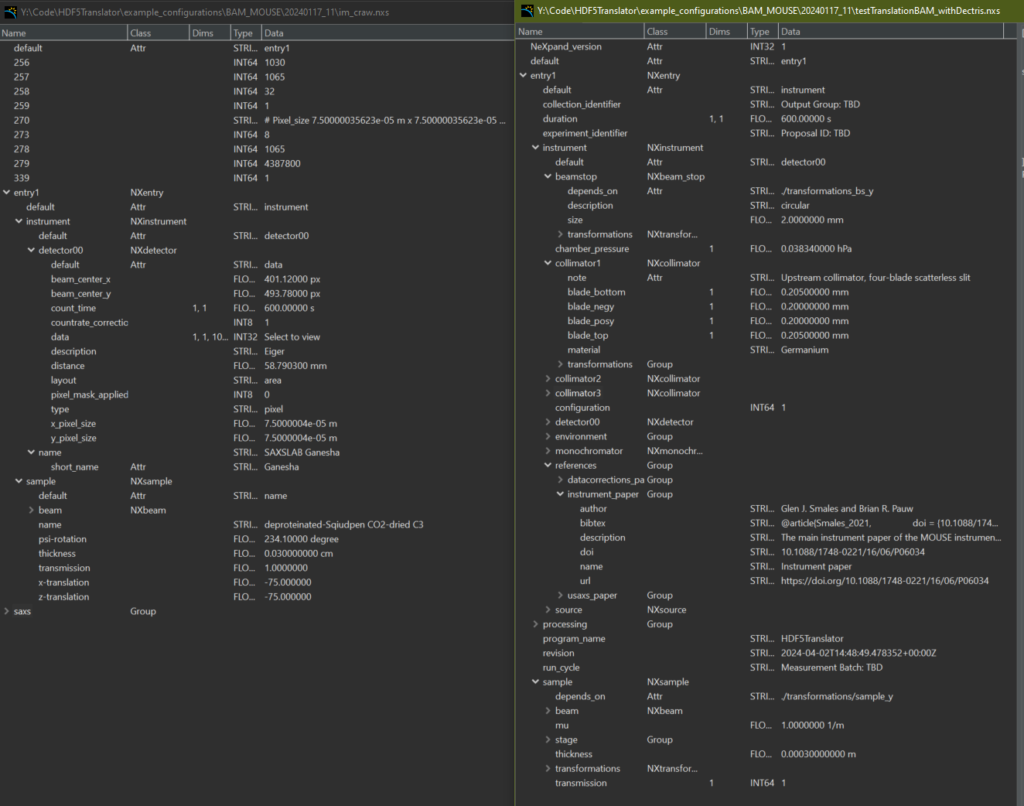
We imagine this tool to be useful across laboratories to homogenize the files in the community. They could be easily used to improve the structure of laboratory data but also synchrotron and neutron source data. As more examples become available, you might just find a ready-to-go translation that would work for you.
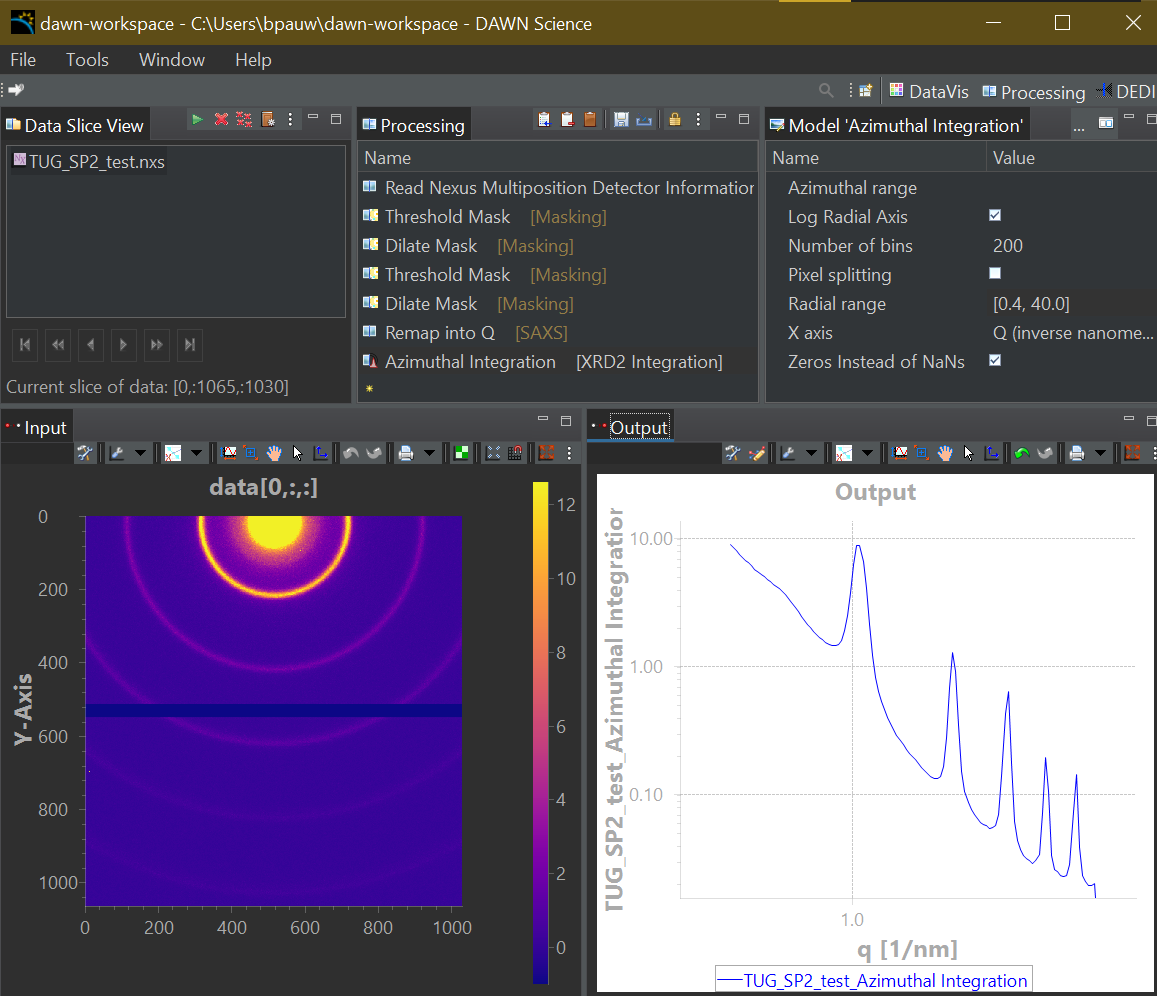
Post-translation processes
As for the post-translation processes, these can be very powerful but are a bit more bespoke. We use ours to determine the beam center, flux and transmission values from the direct beam image with and without sample, and to stack multiple single measurement repetitions into a single multi-repetition measurement file. Likewise you can imagine a ton of useful metadata operations that would help your situation, for example feeding in extra metadata from an electronic logbook into your files. These steps will eventually be separated out of the HDF5Translation package to a separate library of operations. For now, the example is there so you have a flexible framework to modify to suit your needs (the CLI interface takes a measurement file, one or more optional auxiliary files, and keyword-value pairs if you need).
What’s next?
To accelerate its use, we recommend using a sequencing engine such as RunDeck to make it easier for end users to use the tools, or automate them in your data pipeline. If you have a file watcher, you could automatically convert files on the fly as they come off your instrument. The process is reasonably performant and should not impact your data flow.
We hope this package is to your liking, and we hope it will encourage collaboration within the community to extend the translation configuration library. If you have made another example using this framework, do contact us so we can add it to the library. We will continue working on fine-tuning the examples and extending the post-translation operations to carry out the more specific data pipeline tasks. Additionally, work on the post-translation processing steps would also be much appreciated!
